| Anchor | ||||
|---|---|---|---|---|
|
The most recommended way to get the newest version of the deployment recipe is to directly clone the official github repository :
git clone https://github.com/CogStack/CogStack-Recipes.gitThe content will reside in CogStack-Recipes directory.
Alternatively, one can download the bundle ( master branch) directly from the repository:
wget 'https://github.com/CogStack/CogStack-Recipes/archive/master.zip'
unzip master.zip
The content will be decompressed into CogStack-Recipes-master/ directory.
In this tutorial, we will be using the recipes from full-pipeline sub-directory.
| Anchor | ||||
|---|---|---|---|---|
|
The recipe's content
The content of the CogStack recipe bundle (directory: full-pipeline ) consists of such subdirectories:
ingest-documentsnlp-annotaterawdata
The ingest-documents and nlp-annotate directories contain the recipes for deployments of example workflows used for ingesting EHR data with free-text documents followed by the extraction of annotations from the documents. The example EHR data, used as an input for the ingestion pipeline is stored in rawdata directory. The same example data is used in Examples.
CogStack platform
Overview
An example CogStack platform deployment consists of multiple inter-connected microservices running together. For the ease of use and deployment we use Docker (more specifically, Docker Compose), and provide Compose files for configuring and running these microservices. The selection of running microservices depends mostly on the use-case, including specification of EHR data source(s), data extraction, processing and querying requirements.
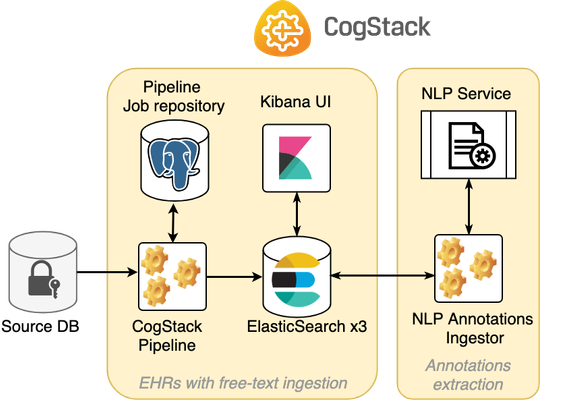
The figure below presents a diagram of an example CogStack platform deployment covered in this tutorial.
The whole EHR data ingestion process is divided into two separate workflows:
- (1) ingestion and harmonisation of EHR with free-text documents data and storing them into ElasticSearch (materials in
ingest-documentsdirectory ), - (2) NLP annotations extraction from the documents ingested in stage (1) with storing the annotations back into ElasticSearch (materials in
nlp-annotatedirectory).
Each workflow uses its own Docker Compose file with the used services definition and configuration.
Available services
The example CogStack platform is composed of the following services (the service names are defined in separate Docker Compose files):
- CogStack-Pipeline – the EHR data primary ingestion pipeline (service:
cogstack-pipeline). - Pipeline Job repository – a PostgreSQL database for storing information about CogStack-Pipeline jobs (service:
cogstack-job-repo). - ElasticSearch – the proposed EHR data store and search engine (3-node deployment) for storing and querying the processed EHR data (service:
elasticsearch-*). - Kibana – the proposed EHR data exploratory and visualisation tool for querying the data residing in ElasticSearch (service:
kibana). - NLP Service – an example NLP application. In our example, used to annotate common drug names (service:
nlp-gate-drug). The application is based on GATE NLP Framework and is running as a service with exposed REST endpoint. - NLP Annotations Ingester – an example annotation ingester application to ingest the documents from ElasticSearch, send to NLP service and send-back to ElasticSearch (service:
annotations-ingester).
Please note that although the Source database (service: samples-db ) has been defined in the Compose file, it only serves here as an example of the input database source (in our case, we use PostgreSQL database) for the CogStack-Pipeline component. The input source for the CogStack-Pipeline usually will be provided as an external database.
Following, we will be analysing the example end-to-end deployment composed of two workflows: documents ingestion and NLP annotations extraction.
Documents ingestion
The documents ingestion workflow content is provided in the ingest-documents directory. The directory contains 2 recipes for the deployment of an example documents ingestion workflow:
- single-step ingestion (directory:
single-step-[raw-text]), - two-step ingestion (directory:
two-step-[documents]; not covered in this tutorial ).
The primary aim of the documents ingestion workflow is to perform the initial ingestion of the documents with data harmonisation from a specified data source to the specified data sink. The data available after the primary ingestion should be ready-to-use by further downstream applications. Usually the data sink would be selected as ElasticSearch and the data can be already browsed and explored in Kibana.
More details about the ingestion workflow are presented in the section below.
| Info | ||
|---|---|---|
| ||
Please note, that for simplicity of this tutorial, we will be mainly covering only single-step documents ingestion workflow, which is based on Example 2. In this example, the free-text data has been already extracted from the documents, originally residing in PDF, Word, images, etc. The two-step documents documents ingestion pipeline is based on Example 5 and we refer the reader to the mentioned example for in-detail description. |
NLP annotations extraction
The nlp-annotate directory contains the recipe for the deployment of an example NLP annotations extraction workflow. The primary aim of the NLP annotations extraction workflow is to provide a set of NLP services and the post-ingestion pipeline component to extract the annotations from the previously ingested documents and store in specified data sink – usually, this would involve reading the documents from and storing the annotations back in ElasticSearch.
More details about the NLP annotations extraction workflow are presented in the section below.
| Info | ||
|---|---|---|
| ||
Please note, that for simplicity of this tutorial, we will be mainly covering the NLP ingestion workflow using a simple NLP application for annotating common drug names as provided in Example 8. The recommend NLP application to run is Bio-Yodie, however it requires the UMLS license to prepare and use the resources and these we cannot include. For more information please refer to Bio-Yodie part of the CogStack documentation. |
| Anchor | ||||
|---|---|---|---|---|
|
General information
In the current implementation, CogStack Pipeline can only ingest EHR data from a specified input database, from a predefined database table / view. This is why, in order to process the sample patient data covered in this tutorial, one needs to create an appropriate database schema and load the data.
Moreover, as relational join statements have a high performance burden for ElasticSearch, the EHR data is best to be stored denormalised and indexed in ElasticSearch. This is why, for the moment, we rely on ingesting the data from additional view(s) created in the sample database. Apart from that, storing denormalised EHRs alongside free-text data allows for performing Google-like queries in Kibana and retrieving full records with the matched keywords.
Following, we cover the process of defining the required schema step-by-step.
| Info | ||
|---|---|---|
| ||
Please note that there is available ready-to-use database dump with the schema already defined and all the sample data loaded – the compressed file is available in the This database dump will be automatically loaded into Source PostgreSQL database ( |
Sample data
The sample dataset used in this tutorial consists of two types of EHR data:
- Synthetic – structured, synthetic EHRs, generated using synthea application (each table stored as CSV file),
- Medial reports – unstructured, medical health report documents obtained from MTSamples.
These datasets, although unrelated, are used together to compose a combined dataset.
For a more detailed description of the dataset used with data samples, please refer to Quickstart tutorial.
Database schema
Tables
Patients table
The patients table definition in PostgreSQL according to the Synthea records specification:
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
CREATE TABLE patients ( id UUID PRIMARY KEY, birthdate DATE NOT NULL, deathdate DATE, ssn VARCHAR(64) NOT NULL, drivers VARCHAR(64), passport VARCHAR(64), prefix VARCHAR(8), first VARCHAR(64) NOT NULL, last VARCHAR(64) NOT NULL, suffix VARCHAR(8), maiden VARCHAR(64), marital CHAR(1), race VARCHAR(64) NOT NULL, ethnicity VARCHAR(64) NOT NULL, gender CHAR(1) NOT NULL, birthplace VARCHAR(64) NOT NULL, address VARCHAR(64) NOT NULL, city VARCHAR(64) NOT NULL, state VARCHAR(64) NOT NULL, zip VARCHAR(64) ) ; |
Encounters table
Similarly, the corresponding encounters table definition:
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
CREATE TABLE encounters ( id UUID PRIMARY KEY NOT NULL, start TIMESTAMP NOT NULL, stop TIMESTAMP, patient UUID REFERENCES patients, code VARCHAR(64) NOT NULL, description VARCHAR(256) NOT NULL, cost REAL NOT NULL, reasoncode VARCHAR(64), reasondescription VARCHAR(256), ) ; |
Observations table
The next table is observations and the corresponding table definition:
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
CREATE TABLE observations ( -- needed for CogStack Pipeline Document data model and ingestion: cid SERIAL PRIMARY KEY, -- > ingestion: primary key created TIMESTAMP DEFAULT CURRENT_TIMESTAMP + RANDOM() * INTERVAL '5 years', -- > ingestion: timestamp document TEXT DEFAULT NULL, -- > nlp: document content (loaded from MTSamples) doc_id INT DEFAULT NULL, -- > nlp: document id date DATE NOT NULL, patient UUID REFERENCES patients, encounter UUID REFERENCES encounters, code VARCHAR(64) NOT NULL, description VARCHAR(256) NOT NULL, value VARCHAR(64) NOT NULL, units VARCHAR(64), type VARCHAR(64) NOT NULL ) ; |
The patient and encounters tables have their primary keys (id field) already defined (of UUID type) and are included in the input CSV files in the generated Synthea files.
However, here have been marked additional document and doc_id fields. These extra fields will be used to store the document content from MTSamples dataset and to be processed by the NLP application.
| Info | ||
|---|---|---|
| ||
Just to clarify, Synthea-based and MTSamples are two unrelated datasets. Here, we are extending the synthetic dataset with the clinical documents from the MTSamples to create a combined one, to be able to perform more advanced queries. |
Moreover, some fields been additionally marked as they have their values auto-generated – these are:
cid– an automatically generated primary key (with unique value),created– a record creation timestamp.
These fields will be later used by CogStack Pipeline for data partitioning when ingesting the records (see creating the observations_view ).
Views for CogStack
Next, we define a observations_view that will be used by CogStack Pipeline ingest the records from the input database:
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
CREATE VIEW observations_view AS SELECT p.id AS patient_id, p.birthdate AS patient_birth_date, p.deathdate AS patient_death_date, p.ssn AS patient_ssn, p.drivers AS patient_drivers, p.passport AS patient_passport, p.prefix AS patient_prefix, p.first AS patient_first_name, p.last AS patient_last_name, p.suffix AS patient_suffix, p.maiden AS patient_maiden, p.marital AS patient_marital, p.race AS patient_race, p.ethnicity AS patient_ethnicity, p.gender AS patient_gender, p.birthplace AS patient_birthplace, p.address AS patient_addr, p.city AS patient_city, p.state AS patient_state, p.zip AS patient_zip, enc.id AS encounter_id, enc.start AS encounter_start, enc.stop AS encounter_stop, enc.code AS encounter_code, enc.description AS encounter_desc, enc.cost AS encounter_cost, enc.reasoncode AS encounter_reason_code, enc.reasondescription AS encounter_reason_desc, -- needed for CogStack Pipeline ingestion partitioner: obs.cid AS observation_id, -- > serves as unique id obs.created AS observation_timestamp, -- > serves as timestamp obs.date AS observation_date, obs.code AS observation_code, obs.description AS observation_desc, obs.value AS observation_value, obs.units AS observation_units, obs.type AS observation_type, obs.document AS document_content, -- will be used for the NLP processing obs.doc_id AS document_id -- (*) FROM patients p, encounters enc, observations obs WHERE enc.patient = p.id AND obs.patient = p.id AND obs.encounter = enc.id ; |
The goal here is to denormalize the database schema for CogStack Pipeline ingestion to ElasticSearch, as the observations table is referencing both the patient and encounters tables by their primary key. In the current implementation, CogStack Pipeline engine cannot perform dynamic joins over the relational data from specific database tables.
Some of the crucial fields required for configuring CogStack Pipeline engine with Document data model have been marked with comments – these are:
observation_id– the unique identifier of the observation record (typically, the primary key),observation_timestamp– the record creation or last update time.
These fields are later used when preparing the configuration file for CogStack data processing workflow.
Moreover, as a hint, two fields in the above view have been marked – they will be used by the NLP application when extracting the annotations (see NLP ingestion workflow):
document_content– this field will contain the free-text document from which the annotations need to be extracted from,document_id– the unique id of the document.
Database schema – the full definition
The full definition of the database schema used is stored in db_prepare/db_create_schema.sql file. This definition file is later used when creating a database dump.
The script db_prepare/prepare_db.sh was used to prepare the database dump.
The sample data used to populate the database resides in rawdata folder in the main directory.
| Anchor | ||||
|---|---|---|---|---|
|
Overview
The main aim of the documents ingestion workflow is to perform the ingestion of EHR data from a specified input source, perform data harmonisation with extraction of the text from the clinical notes and store the processed data in a specified sink. Usually, the sink will be ElasticSearch, serving as the final destination of the ingestion. However, in some cases a database sink can be used, serving as a staging area for the data before ingesting to ElasticSearch.
The EHR data, once already available in ElasticSearch can be already queried using Kibana user interface or directly through the exposed ElasticSearch REST API.
The ingestion is being performed by /wiki/spaces/COGEN/pages/37945560. In the current version, CogStack Pipeline can perform only point-to-point ingestion of EHR data, i.e. reading from a specified database table or view. However, usually a table/view needs to be specially prepared for CogStack Pipeline prior the ingestion as covered in the previous section, to be compatible with CogStack Pipeline Document data model and data partitioner functionality.
There are two workflows available:
- Single-step ingestion (directory:
full-pipeline/),single-step-[raw-text] - Two-step ingestion (directory:
full-pipeline/two-step-[documents]).
In this tutorial, however, we focus only on the single-step ingestion example.
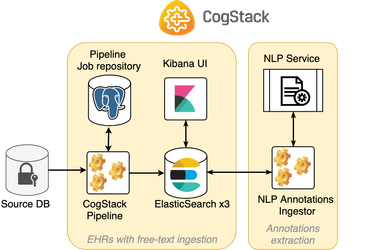
Single-step ingestion
In this example, CogStack Pipeline performs EHR data ingestion where the free-text data is already available in the input database. It does not hence need to run Tika/TesseractOCR on the binary documents to extract the text.
The content is placed in full-pipeline/ directory.single-step-[raw-text]
The diagram presenting a sample deployment based on this workflow is presented below:
This example documents ingestion workflow is based on Example 2.
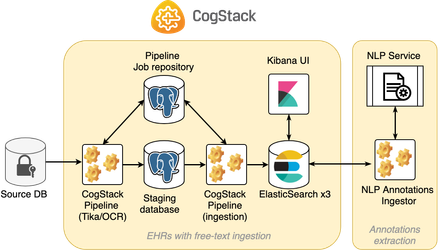
Two-step ingestion
In this example, CogStack Pipeline performs 2-step EHR data ingestion, since the documents are initially stored in binary form and free-text data needs to be firstly extracted. Hence, the workflow is divided into two steps:
- Ingestion of the documents from the input database with performing text extraction from the binary documents using Tika/TesseractOCR. The data is stored in a temporary staging database.
- Ingestion of the EHR with free-text data from staging database into ElasticSearch.
The content is placed in full-pipeline/ directory.two-step-[documents]
The diagram presenting a sample deployment based on this workflow is presented below:
This example documents ingestion workflow is based on Example 5 and will not be covered in this tutorial.
Deployment configuration
Overview
For the ease of use and deployment, the application stack is usually deployed using Docker Compose with each application running as a service. For each application there is a provided docker image with all the necessary prerequisites satisfied. Only the service-specific configuration files and container environment variables may need to be specified prior to running a specific deployment.
The definition of configuration files, Docker Compose files and the running services are covered below.
Services and configuration
Docker Compose file docker-compose.yml with the services definition and configuration has been presented below:
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
version: '3.5'
# need version '3.5' for setting external networks name
#---------------------------------------------------------------------------#
# Used services #
#---------------------------------------------------------------------------#
services:
#---------------------------------------------------------------------------#
# Postgres container with sample data #
#---------------------------------------------------------------------------#
samples-db:
image: postgres:11.1-alpine
volumes:
- ./db_dump:/data/:ro
- ./common/pgsamples/init_db.sh:/docker-entrypoint-initdb.d/init_db.sh:ro
- samples-vol:/var/lib/postgresql/data
ports:
# <host:container> expose the postgres DB to host for debugging purposes
- "5555:5432"
networks:
- default
#---------------------------------------------------------------------------#
# CogStack-Pipeline related containers #
#---------------------------------------------------------------------------#
cogstack-pipeline:
image: cogstacksystems/cogstack-pipeline:dev-latest
environment:
- SERVICES_USED=cogstack-job-repo:5432,samples-db:5432,elasticsearch-1:9200
- LOG_LEVEL=info
- LOG_FILE_NAME=cogstack_job_log
- FILE_LOG_LEVEL=off
volumes:
- ./cogstack:/cogstack/job_config:ro
depends_on:
- cogstack-job-repo
- samples-db
- elasticsearch-1
command: /cogstack/run_pipeline.sh /cogstack/cogstack-*.jar /cogstack/job_config
networks:
- esnet
- cognet
- default
cogstack-job-repo:
image: postgres:11.1-alpine
volumes:
- ./common/pgjobrepo/create_repo.sh:/docker-entrypoint-initdb.d/create_repo.sh:ro
- cogstack-job-vol:/var/lib/postgresql/data
# Job repository should be available only internally for cogstack-pipeline
ports:
- 5432
networks:
- cognet
- esnet
#---------------------------------------------------------------------------#
# Elasticsearch cluster #
#---------------------------------------------------------------------------#
elasticsearch-1:
image: docker.elastic.co/elasticsearch/elasticsearch-oss:6.4.2
environment:
- cluster.name=docker-cluster
- discovery.type=zen
# - bootstrap.memory_lock=true
- "ES_JAVA_OPTS=-Xms512m -Xmx512m"
volumes:
- elasticsearch-vol-1:/usr/share/elasticsearch/data
depends_on:
- cogstack-job-repo
ulimits:
memlock:
soft: -1
hard: -1
ports:
- "9200:9200"
networks:
- esnet
elasticsearch-2:
image: docker.elastic.co/elasticsearch/elasticsearch-oss:6.4.2
environment:
- cluster.name=docker-cluster
- discovery.zen.ping.unicast.hosts=elasticsearch-1
- "ES_JAVA_OPTS=-Xms512m -Xmx512m"
volumes:
- elasticsearch-vol-2:/usr/share/elasticsearch/data
depends_on:
- elasticsearch-1
ulimits:
memlock:
soft: -1
hard: -1
ports:
- 9200
networks:
- esnet
elasticsearch-3:
image: docker.elastic.co/elasticsearch/elasticsearch-oss:6.4.2
environment:
- cluster.name=docker-cluster
- discovery.zen.ping.unicast.hosts=elasticsearch-1
- "ES_JAVA_OPTS=-Xms512m -Xmx512m"
volumes:
- elasticsearch-vol-3:/usr/share/elasticsearch/data
depends_on:
- elasticsearch-1
ulimits:
memlock:
soft: -1
hard: -1
ports:
- 9200
networks:
- esnet
#---------------------------------------------------------------------------#
# Kibana webapp #
#---------------------------------------------------------------------------#
kibana:
image: docker.elastic.co/kibana/kibana-oss:6.4.2
environment:
ELASTICSEARCH_URL: http://elasticsearch-1:9200
volumes:
- ./common/kibana/config/kibana.yml:/usr/share/kibana/config/kibana.yml:ro
depends_on:
- elasticsearch-1
ports:
- "5601:5601"
networks:
- esnet
#---------------------------------------------------------------------------#
# Docker named volumes #
#---------------------------------------------------------------------------#
volumes:
samples-vol:
driver: local
cogstack-job-vol:
driver: local
elasticsearch-vol-1:
driver: local
elasticsearch-vol-2:
driver: local
elasticsearch-vol-3:
driver: local
#---------------------------------------------------------------------------#
# Docker networks. #
#---------------------------------------------------------------------------#
networks:
esnet:
driver: bridge
name: esnet-ext
cognet:
driver: bridge
|
Services
The used services in this docker compose file are:
samples-db– serves as the Source Database. It uses a PostgreSQL database and will load the sample data from a provided database dump (read fromdb_dumpdirectory on the host machine).cogstack-pipeline– the EHR and free-text ingestion pipeline that will run the ingestion job as defined infull-pipeline/single-step-[raw-text]/cogstack/observations.propertiespath.cogstack-job-repo– the job repository database that is used by CogStack Pipeline Spring Batch functionality.elasticsearch-*– ElasticSearch cluster, 3-node deployment,kibana– KIbana user interface that will be used to interact with the data stored in ElasticSearch.
| Info | ||
|---|---|---|
| ||
Please note that the containers with the used services are using the official images either pulled from the cogstacksystems or from the official elastic Docker Hub. However, the specific configuration files for the individual services need to be properly set up for the ingestion process. These files are provided locally and linked with the running containers through path mapping provided in volumes section of the service. |
More details about configuring each of the services are presented in sections below.
Networking
There are two networks specified in the compose file: cognet and esnet .
cognet is an internal network that is used to isolate the running CogStack Pipeline ingestion process with the job repository from other services.
esnet is a network that has been created to isolate ElasticSearch cluster with Kibana from other services. This network will be further exposed as an external network under the name esnet-ext and it will be used by the annotations-ingester during the NLP annotations ingestion step.
Moreover, some of the services bound to the host machine are exposing ports that can be used by external applications:
samples-dbdabatase binds5555port on the host machine to the default PostgreSQL port5432in the container, so one can be able to inspect the input data source,kibanaservice binds5601port on the host machine to the port5601in the container, so the interface can be accessible by external services,elasticsearch-1service binds9200port on the host machine to the port9200in the container, so the external applications can access the ElasticSearch REST endpoint.
Volumes
There are three kinds of volumes used in this example:
samples-vol– the volume that will persist the data fromsamples-dbdatabase,cogstack-job-vol– the volume that will persist the CogStack Pipeline ingestion jobs information,elasticsearch-vol-*– the volumes that will persist the data already ingested in ElasticSearch.
Running the services
One can run the services by using Docker Compose.
To run all the services, with the services configuration stored in the docker-compose.yml file, type in the full-pipeline/single-step-[raw-text] directory:
docker-compose up
Alternatively, one can run only selected services by specifying their names:
docker-compose up <service-name>
| Tip | ||
|---|---|---|
| ||
Please note that this workflow needs to be run prior to running NLP ingestion workflow, which requires access to ElasticSearch cluster and |
| Note | ||
|---|---|---|
| ||
Please note that ElasticSearch uses a sysctl -w vm.max_map_count=262144 For more information please refer to the official ElasticSearch documentation. |
CogStack Pipeline
The data processing workflow of CogStack Pipeline is based on Java Spring Batch framework. Not to dwell too much into technical details and just to give a general idea – the data is being read from a predefined data source, later it follows a number of processing operations with the final result stored in a predefined data sink. CogStack Pipeline implements variety of data processors, data readers and writers with scalability mechanisms that can be selected in CogStack job configuration. The information about the running and finished ingestion jobs is stored in CogStack job repository (usually, a PostgreSQL database). For more details about the CogStack Pipeline functionality, please refer to /wiki/spaces/COGEN/pages/37945560.
When ingesting the EHR data back to the database, appropriate tables need to be created prior to ingestion according to the CogStack Pipeline document data model (for more information please see: /wiki/spaces/COGEN/pages/37945560 documentation and Example 2). However, when ingesting the records to a specified ElasticSearch index, the index mapping will be created dynamically, not requiring any prior interaction.
CogStack Pipeline is running as a service under name cogstack-pipeline .
CogStack Pipeline job repository
CogStack Pipeline ingestion application requires a database that will be used as job repository. In this deployment example, PostgreSQL database is used which is running as cogstack-job-repo service.
CogStack Pipeline ingestion job configuration
Prior running, CogStack Pipeline requires to specify all the ingestion job properties including data source/sink, used components, etc which are specified in the job configuration file. In this example, the ingestion job configuration is specified in cogstack/observations.properties file. The code snippet below presents the configuration with a comment – for a more detailed description of the properties used, please refer to /wiki/spaces/COGEN/pages/37945560 documentation.
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
## ACTIVE SPRING PROFILES ## spring.profiles.active = jdbc_in,elasticsearchRest,localPartitioning #### SOURCE: DB CONFIGURATIONS ## source.JdbcPath = jdbc:postgresql://samples-db:5432/db_samples source.Driver = org.postgresql.Driver source.username = test source.password = test # optional (default: 10): number of allocated connections to source DB (kept until the end of processing) #source.poolSize = 10 # The principle SQL block that specifies data to process. Composed of three parts. source.selectClause = SELECT * source.fromClause = FROM observations_view source.sortKey = observation_id # The principle DB column label mapping for Document data model source.primaryKeyFieldValue = observation_id source.timeStamp = observation_timestamp # Since different DBMS products interpret the SQL standard for time differently, is is necessary to explicitly specify # the date type that the database is using. E.G. postgres=TIMESTAMP, SQL SERVER=DATETIME source.dbmsToJavaSqlTimestampType = TIMESTAMP ##### SINK: ELASTICSEARCH CONFIGURATION ## elasticsearch.cluster.host = elasticsearch-1 elasticsearch.cluster.port = 9200 # optional: store data into this index elasticsearch.index.name = sample_observations_view # optional: if the input SQL query returns columns with these labels, ignore them elasticsearch.excludeFromIndexing = observation_id #### JOB REPO DB CONFIGURATIONS ## jobRepository.JdbcPath = jdbc:postgresql://cogstack-job-repo:5432/cogstack jobRepository.Driver = org.postgresql.Driver jobRepository.username = cogstack jobRepository.password = mysecretpassword #### JOB AND STEP CONFIGURATION ## # optional (default: 50): commit interval in step - process this many rows before committing results. default 50 #step.chunkSize = 50 # optional (default: 5): number of exceptions that can be thrown before job fails. default 5 #step.skipLimit = 5 # optional (default: 2): Asynchonous TaskExecutor Thread pool size - for multithreading partitions #step.concurrencyLimit = 8 #### PARTITIONER CONFIGURATION ## ## This is used to inform how the total row count per job should be broken into ## seperate partitions ## # Two partitioner types are available, either using primary keys (PK) or timestamps and primary keys (PKTimeStamp) # If using the scheduler, the PKTimeStamp type should be configured partitioner.partitionType = PKTimeStamp # optional (default: 1): number of partitions to generate (x / total job row count) #partitioner.gridSize = 1 # name of timestamp column used for partitioning and checking for new data (only if scheduling is used) partitioner.timeStampColumnName = observation_timestamp # name of PK column used for partitioning and checking for new data # only use with scheduling if PKs are guaranteed to be generated sequentially partitioner.pkColumnName = observation_id # this is the table containing the primary keys and optionally, timestamps partitioner.tableToPartition = observations_view ## SCHEDULER CONFIGURATION ## # optional (default: false): if true, run a new job after the last one has finished - new jobs will continue to be created indefinitely #scheduler.useScheduling = false |
Awaiting for the services
Before starting any ingestion job, CogStack Pipeline also will await until specified services are ready – these services are defined in $SERVICES_USED environment variable. These services are used during the ingestion job and have been provided in the job description file.
In this example, the used services variable is defined in the Docker Compose file as:
SERVICES_USED=cogstack-job-repo:5432,samples-db:5432,elasticsearch-1:9200
ElasticSearch and Kibana
ElasticSearch
ElasticSearch is a popular NoSQL search engine based on the Lucene library that provides a distributed full-text search engine storing the data as schema-free JSON documents. Inside CogStack platform it is usually used as a primary data store for processed EHR data by CogStack Pipeline.
Depending on the use-case, the processed EHR data is usually stored in indices as defined in corresponding CogStack Pipeline job description property files (see: CogStack pipeline). Once stored, the data can be easily queried either by using the ElasticSearch REST API (see: ElasticSearch Search API), queried using Kibana or queried using a ElasticSearch connector available in many programming languages.
ElasticSearch apart from standard functionality and features provided in its open-source free version also offers more advanced ones distributed as Elastic Stack (formerly: X-Pack extension) which require license. These include modules for machine learning, alerting, monitoring, security and more.
| Info | ||
|---|---|---|
| ||
Please note, that when ingesting to ElasticSearch without defining the index prior to ingestion, CogStack pipeline will use dynamic mapping. However, to properly use nested data types, ElasticSearch requires prior index definition with proper types. Please refer to ElasticSearch documentation. |
Kibana
Kibana is a data browsing, query and visualisation module for ElasticSearch that be easily used to explore and analyse the data. In sample CogStack platform deployments it can be used as a ready-to-use data exploration and analysis tool.
Apart from providing exploratory data analysis functionality it also offers administrative options over the ElasticSearch data store, such as adding/removing/updating the documents using command line or creating/removing indices. Moreover, custom user dashboards can be created according to use-case requirements. For a more detailed description of the available functionality please refer to the official documentation.
Usage
Using Kibana
In our example, when deploying the platform locally, Kibana dashboard used to query the EHRs can be accessed directly in browser via URL: http://localhost:5601/. The data can be queried using a number of ElasticSearch indices, e.g. sample_observations_view. Usually, each index will correspond to the database view in db_samples (samples-db PostgreSQL database) from which the data was ingested. One can query the data in Kibana by using a rich set of Lucene syntax – please see the official Lucene syntax documentation for more information. Moreover, time series analysis can be performed in Timelion component in Kibana and custom dashboards for data visualisation can be easily created.
| Info | ||
|---|---|---|
| ||
When entering Kibana dashboard for the first time, an index pattern needs to be configured in the Kibana management panel – for more information about its creation, please refer to the official Kibana documentation. |
Using ElasticSearch REST endpoint
In addition, when deploying this example locally, the ElasticSearch REST end-point has been bound to localhost (as specified in Docker Compose file) and so can be accessed via URL http://localhost:9200/. It can be used to perform manual queries or to be used by other external services – for example, one can list the available indices:
curl 'http://localhost:9200/_cat/indices'
or query one of the available indices – sample_observations_view:
curl 'http://localhost:9200/sample_observations_view'
For more information about possible documents querying or modification operations, please refer to the official ElasticSearch documentation.
Using external applications
Since ElasticSearch has a wide community and commercial support, there have been developed connectors for many programming languages. For example, one can easily perform exploratory data analysis in R (see example #1 , example #2).
PostgreSQL (source database)
PostgreSQL is a widely used object-relational database management system. In CogStack platform it is primarily used as a job repository, for storing the jobs execution status of running CogStack Pipeline instances. However, there may be cases where one may need to store the partial results treating PostgreSQL DB either as a data cache (see: Example 5) or an auxiliary data sink.
In our example, we use a PostgreSQL database instance as the data source (apart from a separate instance serving as job repository for CogStack Pipeline). When run, the database will load the generated database dump (full-pipeline/ingest-documents/single-step-[raw-text]/db_dump) with the sample data.
Similarly, in this case, when deployed locally the database port has been bound to localhost (as specified in Docker Compose file) – the PostgreSQL database with the input sample data can be accessed directly at localhost:5555. The database name is db_sample with user test and password test.
To connect to the database, one can run command:
psql -U 'test' -W -d 'db_samples' -h localhost -p 5555
or use DBA tools with user interface, such as PgAdmin.
| Anchor | ||||
|---|---|---|---|---|
|
Overview
The main aim of the NLP ingestion workflow is to allow the extraction the NLP annotations from the free text data, more specifically – from the EHR documents that have been already ingested into ElasticSearch in the previous stage by CogStack Pipeline. The selection of NLP applications is use-case specific and multiple applications can be run in parallel, extracting different types of information from the free-text data.
Since the annotations are ingested-back into ElasticSearch, the annotations can be already browsed and queried in Kibana. Alternatively, the user can manually query the data and build own data extraction and analysis applications by using ElasticSearch REST API endpoint.
All the content used in the ingestion workflow resides in the full-pipeline/nlp-annotate subdirectory.
Deployment
Overview
For the ease of use and deployment, the application stack is usually deployed using Docker Compose, with each application running as a service. For each application there is a provided docker image with all the necessary prerequisites satisfied. Only the application configuration files and container environment variables may need to be specified prior to running a specific deployment.
The definition of configuration files, Docker Compose files and the running services are covered below.
Services and configuration
Docker Compose file docker-compose.yml has been presented below.
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
version: '3.5'
#---------------------------------------------------------------------------#
# Used services #
#---------------------------------------------------------------------------#
services:
#---------------------------------------------------------------------------#
# Ingestion #
#---------------------------------------------------------------------------#
annotations-ingester:
image: cogstacksystems/annotations-ingester:latest
environment:
- SERVICES_USED=elasticsearch-1:9200,elasticsearch-2:9200,elasticsearch-3:9200,nlp-gate-drug:8095
volumes:
- ./pipelines/annotations-ingester/config/config.yml:/app/config/config.yml:ro
command: "bash run.sh"
depends_on:
- nlp-gate-drug
networks:
- nlpnet
- esnet-ext
#---------------------------------------------------------------------------#
# NLP services #
#---------------------------------------------------------------------------#
nlp-gate-drug:
image: cogstacksystems/nlp-rest-service-gate:latest
volumes:
# configuration file
- ./nlp-services/applications/drug-app/config:/app/nlp-service/config:ro
# GATE app resources
- ./nlp-services/applications/drug-app/gate:/gate/app/drug-app:ro
ports:
- "8095:8095"
command: "bash /app/nlp-service/run.sh"
networks:
- nlpnet
#---------------------------------------------------------------------------#
# Docker networks #
#---------------------------------------------------------------------------#
networks:
nlpnet:
driver: bridge
esnet-ext:
external: true
|
Services
The used services in this file are annotations-ingester and nlp-gate-drug . The service containers are using official images from the cogstacksystems Docker Hub. However, the specific configuration files of the individual services need to be properly set up for the ingestion process. These files are provided locally and linked with the running containers through path mapping provided in volumes section of the service.
More details about configuring each of the services are presented below.
Networks
There are two networks specified in the compose file: nlp-net and esnet-ext .
The nlp-net network is an internal network that is used to isolate the running NLP services with the annotations ingester application(s).
The esnet-ext network is an external network that has been defined during the documents ingestion step. It's purpose is to isolate the ElasticSearch nodes with Kibana and possibly with other services that explicitly require access to the ElasticSearch nodes. In this example, only annotation-ingester service require access to the ElasticSearch nodes.
Running the services
One can run the services by using Docker Compose.
To run all the services, with the services configuration stored in the docker-compose.yml file, type in the full-pipeline/nlp-annotate directory:
docker-compose up
Alternatively, one can run only selected services by specifying their names:
docker-compose up <service-name>
| Tip | ||
|---|---|---|
| ||
Please note that this workflow needs to be run after deploying the documents ingestion workflow, as it requires access to ElasticSearch cluster and |
NLP service
Overview
The NLP application used in this is a simple, proof-of-concept application used for annotating common drug names and medications – the application has been already used in Example 8.
The application has been created in GATE NLP suite. GATE NLP suite is a well established set of open-source technologies implementing full-lifecycle solution for text processing.
The NLP application is running as a web service that uses GATE Embedded to run GATE applications. The CogStack NLP REST service wrapper is running as Java (Spring) web application and exposes a REST API endpoint for text processing.
| Info | ||
|---|---|---|
| ||
In the current version, the NLP service only uses applications build in the GATE NLP suite. These applications, e.g. can be previously designed in GATE Developer, exported and configured to be run as an NLP application as a service. |
| Info | ||
|---|---|---|
| ||
Please note that in the current implementation we are using a generic GATE applications runner image and providing application-specific parameters with resources through an external configuration file. The resources with configuration file are present on the host machine and the paths mapped to the docker container in the Docker Compose file. This is why, when running the NLP service for the first time it will require access to the internet to possibly download missing plugins used by the NLP application. The missing plugins will be downloaded from the official Maven repository. |
GATE NLP suite
GATE NLP suite is a well established set of open-source technologies implementing full-lifecycle solution for text processing. Although the GATE ecosystem is very complete, in this tutorial, we only focus on two applications:
GATE Developer is a development environment that provides a rich set of graphical interactive tools for the creation, measurement and maintenance of software components for processing human language. It allows to design, create and run NLP applications using an easy user interface. These applications can be later exported as a custom gapp or xgapp application with the used resources.
GATE Embedded, on the other hand, is an object-oriented framework (or class library) implemented in Java. It is used in all GATE-based systems, and forms the core (non-visual) elements of GATE Developer. In principle, it implements the runtime for executing GATE applications. It allows to run the gapp and xgapp applications that have been previously created in GATE Developer.
NLP drug application
In this tutorial, we use the same application as in Example 8. This simple GATE application can be used to annotate common drugs and medications. The application is using the GATE ANNIE plugin (with default configuration), implementing a custom version of ANNIE Gazetteer. The application was created in GATE Developer studio and exported into gapp format. This application is hence ready to be used by GATE and is stored in full-pipeline/nlp-annotate/applications/drug-app/gate directory as drug.gapp alongside the used resources.
The list of drugs and medications to annotate is based on a publicly available list of FDA-approved drugs and active ingredients. The data can be downloaded directly from Drugs@FDA database. The list of used drugs is stored as drug.lst and active ingredients as active.lst.
More information about creating custom GATE applications can be found in the official GATE documentation.
NLP service configuration
The configuration file for the NLP service ( full-pipeline/nlp-annotate/applications/drug-app/application.properties ) is presented below.
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
# general spring boot configuration
#
server.port = 8095
# Service controller configuration
#
application.class.name = nlp.service.gate.service.GateNlpService
application.name = Example-Drug-App
application.version = 0.1
application.language = ENG
# NLP application specific configuration
#
application.params = { \
gateHome : '/gate/home/', \
gateAppPath : '/gate/app/drug-app/drug.gapp', \
annotationSets : '*:Drug', \
gateControllerNum : 1 \
}
|
The server.port defines the port number within the container which the web service application will be using.
The application.name , application.version and application.language define a general information about the application that will be exposed.
When linking and configuring a specific GATE application with the NLP service, the application.params property need to be defined (represented as key-value pairs, these are application-specific parameters):
gateHome– specifies the path to the GATE Embedded installation dir in the container (this should not be modified, GATE framework is already installed in the service image),gateAppPath– specifies the path to the used GATE application (this path in the container needs to be mapped to a local path on the host through the Docker Compose configuration file),gateControllerNum– the number of processing threads that will be used,annotationSets– (optional) the annotation sets that will be output by the application, can be left empty so all supported annotations by the application will be output.
For a more detailed information about the properties used, please refer to the NLP Service documentation.
| Info | ||
|---|---|---|
| ||
Please note that the paths used in this configuration file correspond to the paths within the container. The paths within the container are mapped to a proper path on the host through the Docker Compose configuration file. This way, a service configuration file is provided with NLP application resources. |
NLP REST API specification
Overview
The NLP service exposes two endpoints:
-
/api/info– provides basic information about the running NLP application (GETmethod ), -
/api/process– used to process the documents and returning the annotations (POSTmethod ).
Details about these endpoints are presented below.
Endpoint: /api/info
This endpoint returns general information about the NLP application that has been set in the configuration file.
It uses GET method to access the resource.
The data returned includes the application-specific configuration that has been provided in the NLP service configuration file. The result is represented in JSON format.
Endpoint: /api/process
This endpoint is used for processing the documents. In brief, the client sends the text to be processed and the service returns the annotations extracted from the text. The type of annotations that will be returned depend on the underlying NLP application used.
It uses POST method to access the resource with sending the payload.
It consumes the data in JSON format embedded in the request body. On success, it returns the annotations with some additional metadata related with the document. The result is represented in JSON format.
Request Body
The request body content, provided in JSON, can be defined as:
| Code Block | ||||||||
|---|---|---|---|---|---|---|---|---|
| ||||||||
{
"content": {
"text": "",
"metadata": {}
},
"application_params" : {},
"footer" : {}
} |
The following objects are available:
content(object; obligatory) – specifies the document content to be processed by the NLP application, where:text(string; obligatory) – contains the free-text data to be processed,metadata(object; optional) – contains additional application-specific information related with the document that will be used by the application,
application_params(object; optional) – additional NLP application-specific parameters to be passed to the processing application (these are NLP application-specific),footer(object; optional) – data that should be returned, included within the response.
Response Body
The response body content, returned in JSON, is defined as:
| Code Block | ||||||||
|---|---|---|---|---|---|---|---|---|
| ||||||||
{
"result": {
"text": "",
"annotations": [],
"metadata": {},
"success": true | false,
"errors": []
},
"footer" : {}
} |
The following fields are available:
result(object, obligatory) – contains the result of processing the text through the NLP application. As the output is application-specific, not all the fields may be output (hence, are optional):text(string) – contains the free-text that was processed or modified during the process (e.g., correction of the sentences),annotations(array) – the annotations extracted from the provided document,metadata(object) – additional, application-specific output related with the processed document,success(boolean) – the indicator specifying whether the processing was successful (true) or there was an error (false),errors(array) – a list of possible errors that occurred during the processing.
footer(object; optional) – the copy of the data that has been provided in footer in the request body.
Annotations ingester
Overview
The annotations ingester is a simple Python application that implements the ingestion and extraction of NLP annotations from documents from/to ElasticSearch. The processing steps are implemented as follows:
- ingester reads the documents from a specified ElasticSearch source index,
- sends the free-text content from a selected field to a specified NLP service,
- receives back the annotations and stores them in a specified ElasticSearch sink index.
All the parameters describing the data source, sink, fields mapping with the used NLP service are specified in the appropriate configuration file ( full-pipeline/nlp-annotate/pipelines/annotations-ingester/config/config.yml ).
| Info | ||
|---|---|---|
| ||
Please note that in the current implementation we are using a generic annotations ingester image and are providing application-specific parameters through an external configuration file. This configuration file resides on the host machine and is mapped to the docker container through |
| Info | ||
|---|---|---|
| ||
It needs to be noted that annotations ingester is a simple application and, when run, performs a single ingestion operation. In contrast to CogStack Pipeline it does not implement any automated scheduling nor batch processing – this functionality needs to be handled by the user. We are upgrading handling and defining flows of data in the platform and so this ingestion process will probably change soon. |
Configuration
Ingestion configuration
The configuration file defines the necessary parameters for the ingestion process and is stored in full-pipeline/nlp-annotate/pipelines/annotations-ingester/config/config.yml . The configuration file represented in YAML format.
Below is presented the used configuration file in this example:
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
source:
es:
hosts: ["elasticsearch-1:9200", "elasticsearch-2:9200", "elasticsearch-3:9200"]
index-name: 'sample_observations_view'
sink:
es:
hosts: ["elasticsearch-1:9200", "elasticsearch-2:9200", "elasticsearch-3:9200"]
index-name: 'new_atomic_annotations'
nlp-service:
endpoint-url: 'http://nlp-gate-drug:8095/api/process'
mapping:
source:
text-field: 'document_content'
docid-field: 'document_id'
persist-fields:
- 'encounter_id'
- 'patient_id'
- 'observation_id'
- 'encounter_start'
- 'encounter_end'
batch:
date-field: 'encounter_start'
date-format: 'yyyy-MM-dd'
date-start: '2010-01-01'
date-end: '2018-06-01'
sink:
split-index-by-field: 'type' |
The essential parameters are:
sourceandsink– defines ElasticSearch endpoints with indices to read from/write to the data,nlp-service– defines the NLP service endpoint used to process the free-text data,mapping– defines the mapping of the extracted annotations from free-text document content into the output annotations index (or a set of indices), with the sub-keys:source– relates to documents stored in ElasticSearch source, such as the name of the free-text field with document content (text-field), the unique document id (docid-field) and the list of fields that will be persisted from the document within the annotations (persist-fields).sink– relates to the how the annotations will be stored in the sink, e.g.split-index-by-fielddenotes the name of the field in the input document which will be used to split the index name (optional).
For a more detailed description of the available parameters, please refer to the annotations-ingester documentation.
Awaiting for the services
Before starting any ingestion job, the application also will await until specified services are ready – these services are defined in $SERVICES_USED environment variable. These services are used during the ingestion job and have been provided in the job configuration file.
In this example, the used services variable is defined in the Docker Compose file as:
SERVICES_USED=elasticsearch-1:9200,elasticsearch-2:9200,elasticsearch-3:9200,nlp-gate-drug:8095
Output format
Each of the extracted annotation will be stored as a separate document within specified index in the ElasticSearch sink (in this example: new_atomic_annotations-* indices). Each annotation will contain a variable number of fields that are prefixed with:
nlp.– the annotations features output by the NLP application,meta.– the selected fields coming from the input document that were selected to be persisted (persist-fieldsin the annotations-ingester configuration file).
The annotations can be linked back to the original document thanks to docid-field specified in the annotations ingester configuration file. Moreover, they can be linked with any other relevant entities by using the specific persisted field (such as patient_id , observation_id , etc.).
| Info | ||
|---|---|---|
| ||
It needs to be noted that the exact number fields with the field names that will be available in each of the annotations depend on:
|
End-to-end data mapping
Primary documents ingestion
In this tutorial, the EHR data with free-text documents are read from samples-db database from observations_view . CogStack Pipeline performs point-to-point ingestion of the records from the specified view where data is represented in a denormalised way.
More specifically, CogStack Pipeline ingests the records using the SQL query: SELECT * FROM observations_view ORDER BY observations_id . This is defined in full-pipeline/ingest-documents/single-step-[raw-text]/cogstack/observations.properties file in the lines:
source.selectClause = SELECT *
source.fromClause = FROM observations_view
source.sortKey = observation_id
The records are ingested into ElasticSearch and stored in sample_observations_view index.
There can be many CogStack Pipeline jobs running in parallel, but each needs to store the data in separate index.
NLP annotations ingestion
In the ingested EHR data with documents, the free-text content is stored in document_content field. The annotations-ingester application reads the content from this field, sends to the NLP application running as a service and receives back the annotations. Each of the received annotations is stored as a separate document in ElasticSearch under the index name new_atomic_annotations-* . There will be multiple indices created, where the suffix of the index will correspond to the value of the field type (as specified in mapping/split-index-by-field property) in the input document.
The annotations stored in ElasticSearch will contain a number of fields prefixed with:
nlp.– annotation features returned from the NLP application,meta.– additional metadata fields that have been persisted (specified inmapping/source/persist-fieldsproperty).
The configuration file for the ingester is stored in full-pipeline/nlp-annotate/pipelines/annotations-ingester/config/config.yml .